Just as there are very common names (e.g., Müller and Schmitt) and rare names, there are haplotypes (a set of results found on the Y chromosome) with a high frequency (also known as common ones) and haplotypes with a low frequency. The 12-67 (or more) STR markers (called markers for short) resulting from the Y-chromosome test are referred to as a haplotype, which can be used to determine whether your DNA sample is common or uncommon.
If you compare a 12-marker result with another 12-marker result from someone with the same surname and results, there is a 99% probability that you are both related within the time frame included in the MRCA (time to a common ancestor in years or generations) tables. If the result is 11 to 12, there is still a high probability that you are related if the 11/12 result is within the same surname.
If you compare a 25-marker result with another 25-marker result with the same surname and the results are 25/25, there is also a 99% certainty that the two people are related, but within a much closer time interval than with the 12-marker test.
If you compare the 12-marker result with someone else who doesn’t have the same surname, but the results match, you’re probably not “recently” related. When we use the term “recently” we’re talking about a timeframe, the last 1,000 years or 40 generations, a time, a depth, in which the earliest known use of surnames lies.
According to current theories, we are all related to one another. The degree of kinship depends only on the timeframe or the number of generations between the subjects and the common ancestor.
We all descend from a single person, as do DNA tests, but what we do is fail to tell ourselves or acknowledge this obvious fact.
Since we all descend from one person and then from a few families, there are naturally always branches through the families, up to the point where we develop our own family nest. It would make sense for us to test our DNA. The fewer markers we test, the less unique we are, and the more markers we test, the more unique the entire set of markers becomes. In other words, pushing the boundaries. If we test just one marker, we would certainly be related to millions of people who have shared these markers for thousands of years.
But on the other hand, if we test a large number of markers, we will find very few people who have the same marker matches as us. These would then be the people who are closely related to us.
This applies when testing our results for 12, 25, or 37 markers. The probability that we will match with other people with 12 markers is far higher than with 25 or 37 markers. This is especially true if our family comes from a population group that originated from one or a few less productive families thousands of years ago (which is the case in Western Europe). Dr. Luigi Cavalli-Sforza Lucca, professor emeritus at Stanford University, writes in his fascinating book, The Great Human Diaspora: The Story of Diversity and Evolution, that the total population of Europe originated from about 60,000 people at the end of the last Ice Age, about 10,000 years ago. Now, Europe has a population of 300 million people. This increase is due almost entirely to natural population increase rather than immigration from other continents. With this in mind, it’s important to note that many people living in Europe today share common ancestry with other Europeans, predating the time when our ancestors began adopting family names, and your first thought for someone with a different surname should always be that the connection is more distant than close.
Our bodies function like copy machines when it comes to Y-DNA. You can have a copier make 1,000 copies without a problem, and then the 1,001st will have an “o” that looks more like an “e.” If we use that copy to make more, all the new ones will have an “e” instead of an “o.” This is a simple way to explain how mutations occur in Y-DNA when it is passed (copied) from father to son. Mutations aren’t common. In fact, they’re very rare, but they can happen randomly over time, meaning I inherited a mutation from my father. This is why all those matches near 12 markers would be lost in most cases if they happened between different surnames. So we increased the numbered markers to compare, because this reveals more mutations, and thus shows us which path leads back to the time when the common ancestor lived.
The only exceptions are if adoption or mistaken paternity has occurred, but this is difficult to prove, though certainly not impossible.
If two 12-marker results match for two subjects with the same surname and the genealogy shows a common ancestor in 1835, the DNA test has confirmed the research and proven that the two descendants are related. In this example, you have two pieces of evidence to support the fact that the tested individuals are related.
First: a documented paper trail, and second: the DNA results. Furthermore, the research provides a precise timeframe for the common ancestor.
(The first 12 STR markers are! These are also used for forensic purposes. Fingerprint!! DYS 19, DYS 390, DYS 391, DYS 392, DYS 393, DYS 3851/II, DYS 3891, and DYS 3851. The last two are duplicative, 2 alleles.)
Without genealogical research, and where two subjects with the same surname match the 12-marker test, the scientific answer regarding the degree of relatedness is that the common ancestor should have lived within 50% of the time of 7 generations, or within approximately 150 years.
The range of generations for the common ancestor extends to 76.9 generations, or almost 2000 years, in cases where there is no common surname. Therefore, the meaning of the surname is of utmost importance in providing a convenient conclusion to the family relationship. For most of the time, when people with different surnames have a random match, a DNA test with a higher number of markers is not useful.
MRCA = Time to the most recent common ancestor/number of generations. (The more markers tested, the higher the price.)
MRCA* tables show you the general probabilities for family relationships at different levels.
DNA sequencing = Marker analysis of the genome (This is generally called a DNA test.)
To explain:
Human DNA[1] is located on 23 chromosomes and consists of 3.4 billion base pairs, each consisting of 4 bases (adenine, guanine, cytosine, thymine).
A deviation from the sequence at one position (e.g., G instead of T) is called a SNP (single-nucleotide polymorphism). This is a single deviation. SNPs occur more frequently in certain sections of DNA, so for genealogical purposes, only those specific sections, where deviations are more likely to occur, are tested. Currently, approximately 650,000 SNPs out of a total of 3.4 billion.
Matching: Comparison (of the autosomal DNA of father and mother) for common sections. Anyone who has had their autosomal DNA tested can upload it anonymously (as so-called raw data) to various databases. The purpose of this is to find possible relatives. The larger (longer) identical sections found in the databases, the closer the possible relatives or common ancestors are.
On average, every 5,230 base pairs are tested, one position; all base pairs in between are not tested. Because SNPs are distributed irregularly in DNA (only in certain sections), the SNPs are actually determined in close succession in some regions of the DNA, and in others in greater succession.
A DNA segment of one centimorgan (cM)[2] in length has approximately 1 million base pairs, so on average, almost 200 SNPs are tested per cM. However, depending on the analytical chip used, not all test providers test the same ones.
The atDNA test for matching purposes, for comparison with the atDNA of all tested subjects, is currently the most commonly used test.
STR = Short Tandem Repeats: Short, consecutive, repeating segments of DNA (this is the explanation for STR markers).
STRs are short DNA sequences a few base pairs long that are repeated multiple times in direct succession.
For this purpose, a Y-DNA test tests for specific markers and specifies the number of their repetitions. STRs vary greatly from person to person (genetic fingerprint)!
SNP = Single Nucleotide Polymorphism. Single-nucleotide gene variant: A SNP (or ‘snip’ in laboratory jargon) refers to a variation of a single base pair in a DNA strand. SNPs are inherited and heritable genetic variants. Low mutation rates over multiple generations. (Particularly suitable for determining descendants or common ancestors! These locations in the DNA strand each mark a haplogroup or haplosubgroup.)
Triangulation, along with phasing, is a method in DNA genealogy for identifying DNA matches over the of the last 300-400 years (back about 10 generations) that prove genetic descent (IBD = Identical by Descent) from a common ancestor. Simply put, triangulation is the creation of triangles.
How do you perform triangulation?
Procedure
This requires at least two independent DNA matches that match on an overlapping part of one of their chromosomes. They should not be more closely related than third cousins.
An example:
Anton (A = the test subject) has a match with Bert (B) on chromosome 15 from position 83,561,856 – 154,688,278. Anton has a match with Charles (C) on chromosome 15 from position 83,752,670 – 154,691,672.
A -> B and A -> C are the first two sides of the triangle; now you have to prove the third side of the triangle. To do this, you check whether Bert (B) also matches Charles (C) on an overlapping position on chromosome 15 (in this example, otherwise any other chromosome).
We find a match between Bert and Charles on chromosome 15 from 88,470,251 to 116,916,652.
Result
This also satisfies the third side of the triangle, and with a very high probability, all three share a common genetic ancestor on the overlapping segment from position 88,470,251 to 116,916,652. This probability increases by phasing individual matches (for example, Anton, if one of his parents also performed a DNA test) or by other DNA cousins who also share a match with Anton, Bert, and Charles on an overlapping position from 88,470,251 to 116,916,652.
Further Notes
The size of the overlapping segment provides an indication of how far back the common ancestor should be sought. The smaller the length, the further back the formula is.
It is preferable to perform both triangulation and phasing on individual matches.
Only 1.5% of the genome is human DNA, which is approximately 30,000 to 35,000 protein-coding genes. The 3 billion base pairs make up the difference between two people.
X gene, 2 XX gene is female. – Y gene, XY is male.
In autosomal DNA testing, 20,000 genes (22 chromosome pairs) must be tested. Cousins up to the 10th degree or across 7 or more generations can be found.
Here’s an example of our approach:
Prerequisite: Objective (Where do our ancestors Forker come from?)
Based on some evidence from our family research, which has been ongoing since 1928, I assumed that our ancestors came from Scotland, from a time before 1300. I wanted to determine this using other namesakes. DNA tests have been available for this purpose since around 2000.
I’ve been exploring the topic of DNA testing since 2009, and in 2011, the Swiss company IGENEA offered me the opportunity to take an “affordable” DNA test. One haplogroup and 12 STR markers were identified. However, this result was still completely insufficient to provide proof of our hypothesis.
This only allows us to determine where (on which continent) male ancestors with the same haplogroup lived several thousand years ago.
I would like to add that over the past few years, the findings have become and continue to become increasingly extensive and differentiated!!
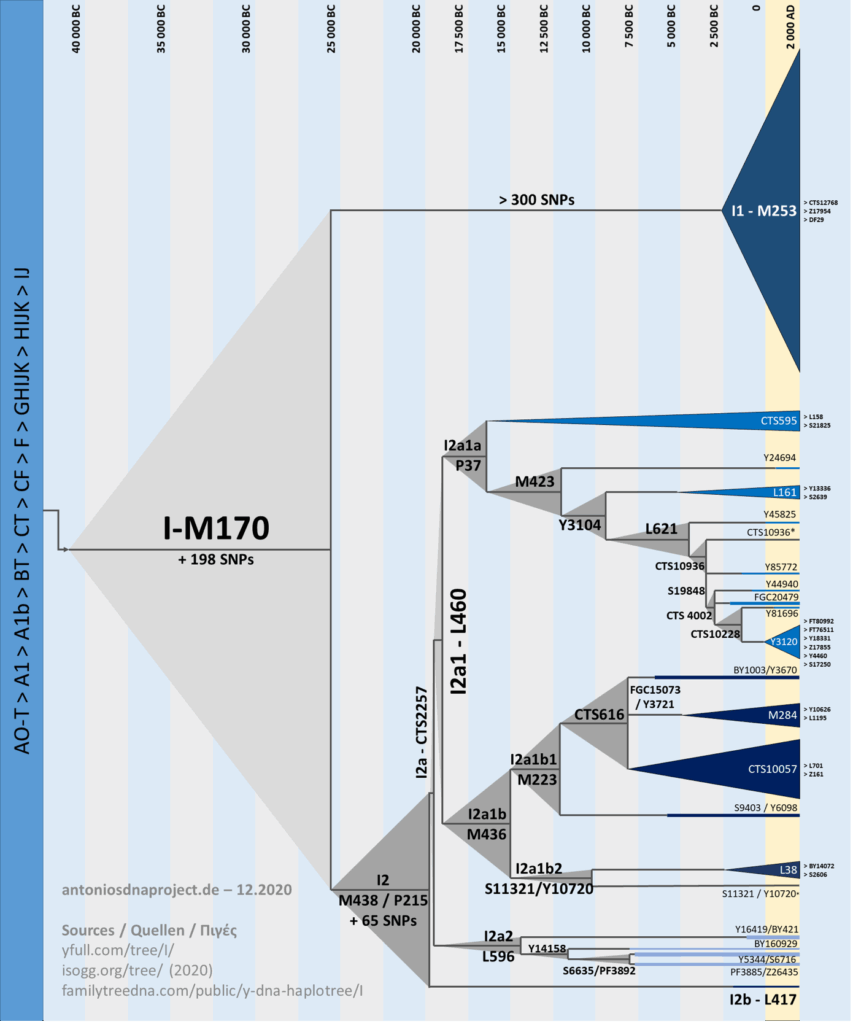
When a Y-DNA test results in a haplogroup, it is crucial where in the haplogroup tree the identified haplogroup is located. The most meaningful is the ultimate one! The one at the end of the tree. (The leaf). You could say it’s the house number.
The system for identifying haplogroups consists of the name of the main group, from A to Z, and a number of subgroups, consecutively numbered with numbers and letters, and finally, an identification of the testing laboratory. In our case, haplogroup I2a and a further 34 subgroups/subgroups up to the currently ultimate subgroup. Whenever another test subject with the same mutation is found in a subgroup in two independent testing laboratories, another haplosubgroup is formed.
This meant first determining the ultimate/final common subgroup of our male name bearers, which, if possible, has only formed in the last hundred years. Because it is known that another (younger) mutation arises approximately every 1200 years.
With our namesake Wolfgang, we tested each other for known haplosubgroups over the course of several years using the offered Aray chips to minimize costs.
The common haplogroup of the Forkers only emerged after several test subjects were tested. From our research (paper research, data from church records), we can prove over 16 generations how the filiation took place, whereby it is known where the surname was passed on through adoption or where the daughter passed on her surname. But ultimately, it turned out that Forker descendants from lines in nearby towns, whose relationship cannot be determined from the earliest church records, are carriers of a common Y-DNA.
Now we are looking for one or more people worldwide with our family name who have also determined their ultimate Y-DNA haplogroup. However, it must also be known where their earliest ancestors lived. Or where DNA archaeologists have found identical DNA from the same haplosubgroup. From what is known to date, our haplogroup points to southern Sweden in one case. Far too few people have been tested yet to find relatives. But at least a lot has been achieved. Our haplosubgroup is not yet in the databases.
Question: Which is more reliable for determining a common ancestor?
The agreement across 67 Y-STR marker positions or is it the ultimate haplogroup (I2-BY135483) as of May 2025. New, unique haplosubgroup.
It is clearly the ultimate Y-DNA subgroup, as this represents the link over significantly longer periods of time. While the proportion of identical Y-STR markers only proves a probable relationship over a few generations.
See Willibald Forker and Wolfgang Furkert, same number of DYS markers (match 37 of 37), same ancestor 12 generations ago (7 generations ago from Lauterbach to Stürza, with a name shift from Forker to Furkert).
Same subgroup and yet different surnames. (Adopted?)
Same surname and yet different subgroup (mother’s name, in our opinion).
One of the largest forums among several others is https://www.ancestry.de/ (original in English *.com). The personal subgroup and the earliest identified ancestor can be transferred to their databases. These data banks are sorted by STR markers, thus making it possible to determine whether there is a relationship. It is still not proven who an unknown name bearer actually underwent testing, since when and where. So, from a time before the data from church records, let’s say around 1200, i.e., from the time before family names were established. Only locally found DNA from skeletons can we determine more precisely where ancestors lived in “more recent times.”
Sources: Igenea, Ancestry, Wikipedia, Gen-Wiki, Yseq Thomas Krahn Berlin.
Summary:
For an initial DNA haplogroup test, you can use the “Y-DNA test with specific arrays” to determine your haplogroup based on a few known haplogroup markers. A simple Y-DNA test only determines which haplogroup and a few subgroups the sample contains.
The ultimate lowest haplosubgroup marker, however, can only be determined in several steps once it is known. (As previously described.) However, if this has already been identified through testing, a close relative of the name bearer only needs to take a test for this ultimate marker, currently 19 euros, T. Krahn. The result is yes or no.
However, to learn about all of their DNA relevant to genealogy (Y-DNA, mt-DNA, and autosomal DNA), the only option is the WGS test. This also reveals previously unknown mutations. Unknown ultimate haplogroup markers can only be determined with a WGS test. Whole genome sequencing (WGS). With the introduction of WGS sequencing, the analysis of the entire human genome (Whole Genome Sequencing, WGS) can be obtained in a single test at a time. Note: with an inexpensive DNA test, only the Y-DNA main group is determined. With inexpensive tests and the aid of electronic arrays (analytic chips), each with approximately four haplogroups (lengthwise or crosswise in the haplogroup tree), the currently known haplogroup can be determined in several steps. Only the WGS test can reveal whether there are further unknown mutations.
Based on the earliest human bones found by archaeologists to date and the DNA extracted from them, this was assigned to haplogroup main group A. (Dating from a time 120,000-156,000 years ago.) Over time, more and more skeletons were examined for their DNA, and it was discovered that the younger the bones, the more changes (mutations) were found. To organize these findings, it was agreed to assign letters from A to Z to each more recent mutation. These denote the main groups, while numbers and lowercase letters are used for their subgroups.
For comparison: we classify trees and simply label tree species from A to Z (spruce, oak, linden, etc.). These are the haplogroups, but we wanted to be more precise, as there are still numerous typical characteristics (mutations) within the haplogroups; these are the haplosubgroups. We’ve only been to the tree trunks. Now we’re moving on to the branches, further haplosubgroups! We want to go down to every single leaf, which is the family name!!! So, via small branches and twigs, down to the last leaf. A long but now well-known path of the last 20 years. It only works via Y-DNA, which is passed on from father to son. Women don’t inherit a Y chromosome from their father; otherwise, they would be sons. You (women) can only access the test for your father’s origins or his DNA through a male descendant of your grandfather, father (or uncle or his son, cousin). This test for the ultimate haplogroup (for the leaf) can be achieved through various methods (tests). An explanation: it has now been recognized that a mutation occurs in the inheritance of a haplogroup approximately every 1200 years. This means that, based on the time of death centuries ago from ancient grave bone finds, one can chronologically determine when men with the mutations present at that time lived in the area. Based on the locations of the finds and the subsequent, younger mutations that have developed, the distribution of this mutation or haplogroup can be graphically represented on Google Maps (world map). In our case, Forker Haplogroup I-BY135483, there are 34 positions (haplosubgroups).
On the way from Africa! See, for example: https://phylogeographer.com/mygrations/?hg=I2&clade=I-BY135483
[1] See GENWIKI
[2] Unit of measurement for 1 million base pairs